mRNABench: a benchmarking suite for property prediction tasks
New tools to advance RNA modeling and benchmark progress in mRNA biology.


TLDR: we introduce a new benchmarking suite for mRNA property prediction tasks. It includes 10 curated datasets and 59 tasks spanning half-life prediction, translation efficiency, RBP binding, localization, and variant effect prediction. Read the paper, check out the mRNABench GitHub, and get started with our easy-to-use API today!
In recent years, mRNA has become an increasingly important modality across a range of therapeutic areas. From vaccines and cytokine therapies to personalized cancer treatments, the number of mRNA-based clinical candidates continues to grow rapidly. Moderna and Merck’s mRNA-4157, for example, has shown durable clinical benefit in melanoma. BioNTech’s recent acquisition of CureVac and AbbVie's acquistion of Capstan Therapeutics, underscores the strategic value of this modality for pipeline expansion. As mRNA transitions from a proof-of-concept to a programmable therapeutic backbone, our ability to design and reason about RNA sequences becomes increasingly important.
Yet despite this shift, our modeling tools remain relatively underdeveloped. Existing nucleotide foundation models are typically trained on DNA or ncRNA sequences and often struggle to capture the biological nuances specific to mature mRNA, such as UTR-mediated regulation, translation dynamics, and isoform-specific function. We believe that mature mRNA deserves dedicated modeling infrastructure.
To address these challenges, we introduce two resources:
- mRNABench: a comprehensive benchmark suite for mRNA property prediction tasks
- A study on how to build more efficient and powerful mRNA foundation models
mRNABench: A Benchmark for mRNA Property Prediction
We’re introducing mRNABench, a benchmark designed to rigorously evaluate models on mature mRNA biology. It includes:
- 10 curated datasets and 59 tasks spanning half-life prediction, translation efficiency, RBP binding, localization, and variant effect prediction
- Three biologically informed data splits (homology, k-mer, and chromosome-based), designed to test generalization across distinct types of sequence variation
- An extensible framework that allows community contributions of new models and datasets.
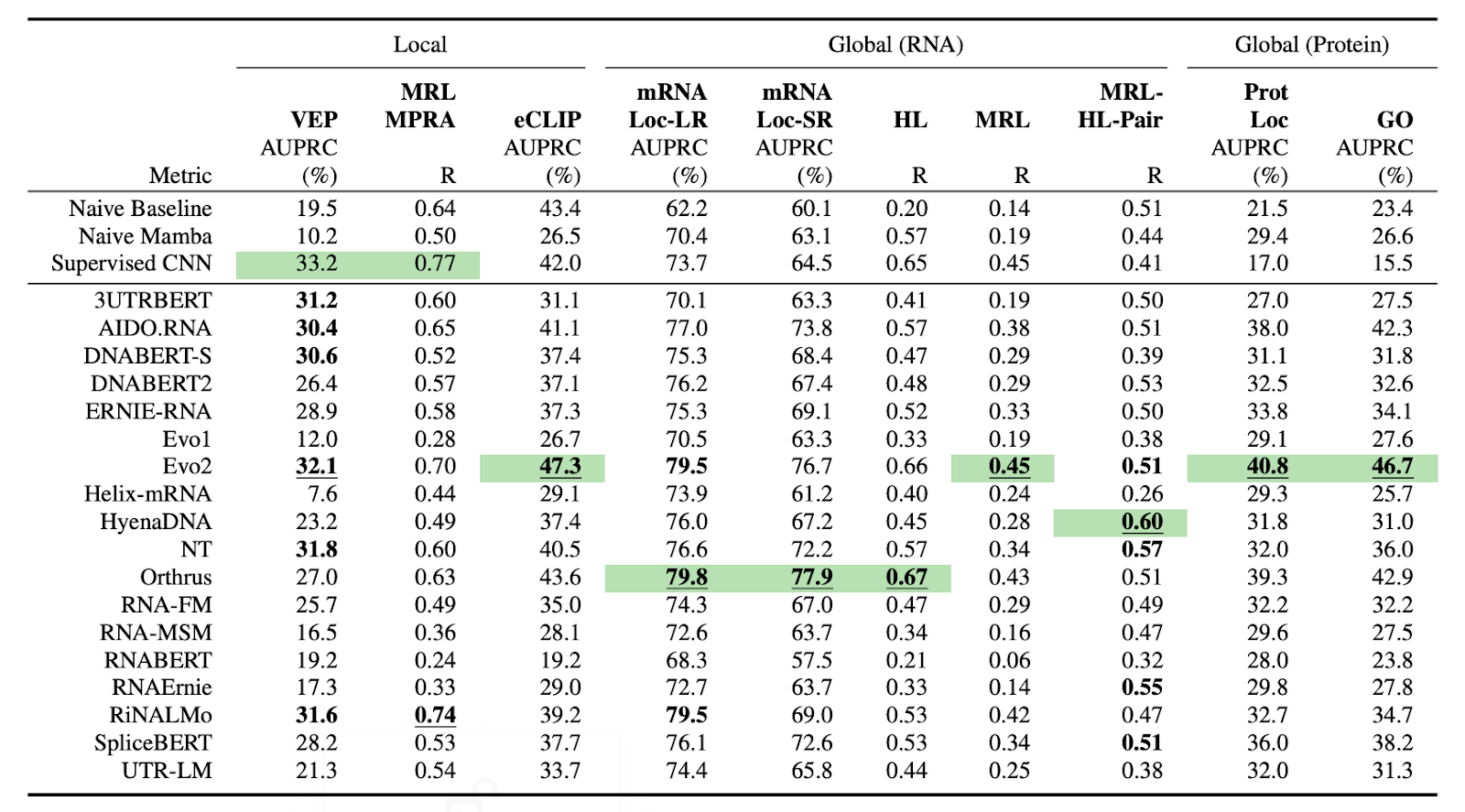
The benchmark also comes with a GitHub repo that provides standardized APIs and evaluation scripts so you can easily load datasets and evaluate your model. Leaderboard coming soon 👀
Get started with just a few lines of code!
Orthrus+MLM: Improving Performance by Combining Training Objectives
Our work on Orthrus+MLM builds on Orthrus, which uses contrastive learning (CL) to capture global relationships between splice isoforms and orthologs. We hypothesized that its performance could be improved by integrating an objective better suited for local sequence features.
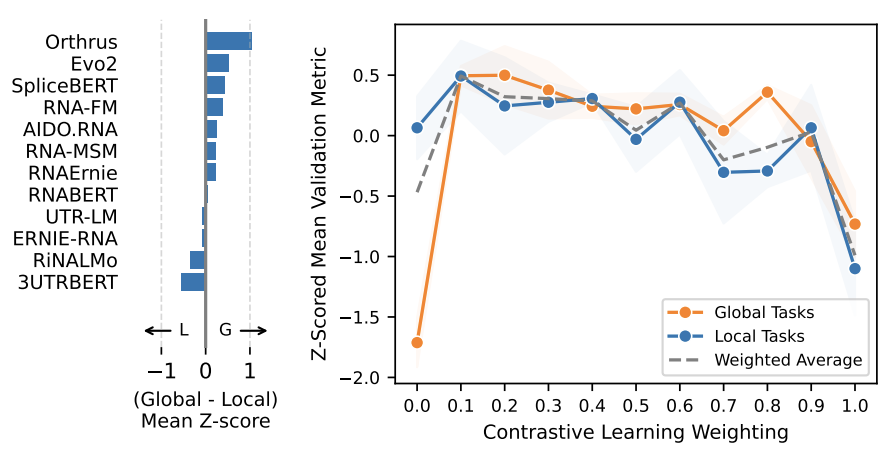
To test this, we introduced a dual-objective framework that combines the global CL strategy from Orthrus with a masked language modeling (MLM) loss. We trained a highly compact Mamba-based architecture (~10 million parameters) using this hybrid approach. Our experiments on mRNABench revealed that adding the local MLM objective improved performance on local tasks and, in addition, significantly boosted performance on global tasks.
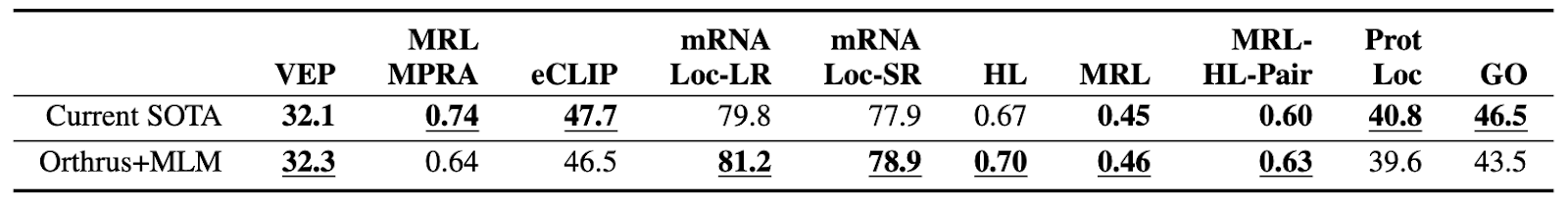
The resulting model, Orthrus+MLM, achieves state-of-the-art performance across the benchmark. Despite its small size, it matches or exceeds the performance of models over 700 times larger.
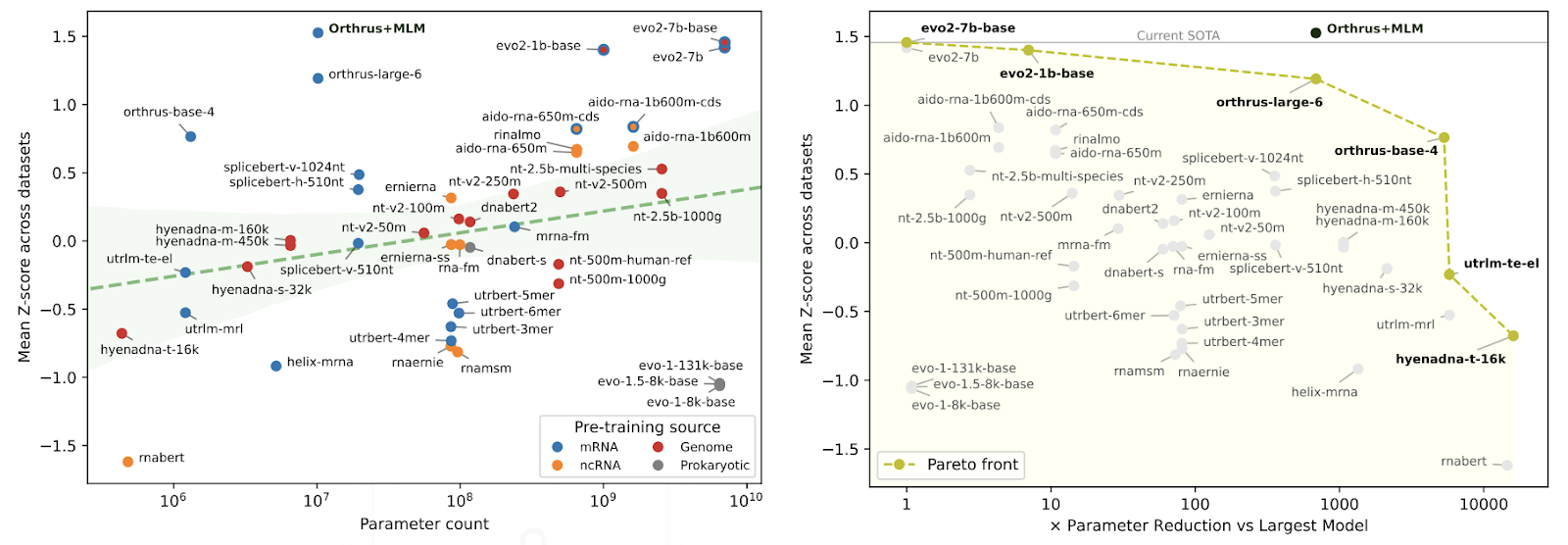
Orthrus+MLM is trained entirely on mature transcript sequences and produces fixed-length embeddings that can be used out-of-the-box for downstream tasks or fine-tuned when additional data is available.
Why These Tools Matter
As the field of mRNA therapeutics matures, there is a growing need for models that can make precise, data-efficient predictions about the effects of sequence variation — whether the goal is to design better UTRs, reduce immunogenicity, or optimize expression in a specific cell type. With mRNABench, we offer a rigorous framework to measure whether models are actually improving on the problems that matter.
Explore the resources today:
We look forward to seeing how the community builds on this work, and we welcome collaborations aimed at expanding both the modeling and benchmarking toolkits for RNA biology.
Learn more about our foundation models
.png)

